NGYX I.C.
WEB Content
NGYX I.C. Validation and Challenge.
This page gives information about NGYX I.C. approach in terms of Validation of a Drug Resistance Prediction System (that also helps to understand how our datasets/subsets were created; Section I) and about an open Challenge (see Section II).
I. NGYX I.C. Validation Methodology.
Remark: Some "rules" may look applicable only to Drug Resistance Prediction System Validation.
Rule 1. Always use 50% of the dataset as "unseen data" for the final validation.
Rule 2. Always find a way to provide a level of confidence of the final validation results/outcomes and question yourself about it and putative "collateral damages". e.g. "Is 5% error good enough or do we need 1% level?"
Rule 3. Identify Methodologies that are applicable for ALL of similar purposes. Do not introduce "variation" into the methodology itself. Stay consistent!
Rule 4. Be always in a position to evaluate what are the maximal and minimal levels of accuracy that can be expected and where your approach lies in between these 2 extremes.
Rule 5. Be Innovative/Creative. And do not take as fact what everyone uses. Just think and create. Believe in you!
Rule 6. Develop a system that is not a "Black Box" (e.g. Neural Networks). Your system should be "informative". e.g. "We use these Mutations to derive Drug Resistance". Otherwise you will be into trouble to validate it on e.g. a Clinical Outcomes Datasets.
Starting with theses rules in mind here is how NGYX I.C. works these out.
Example given: A Geno/Pheno(s) Dataset of 800 Samples (200 with repeated measurements).
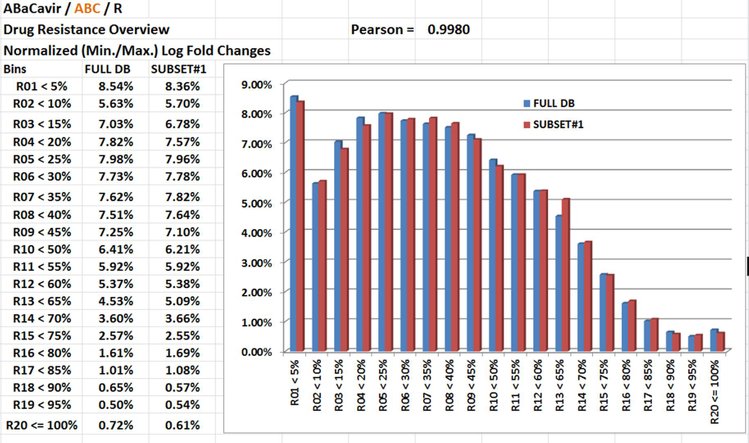
NGYX I.C. first split into four subsets (+/- equal in size ~ 250) the dataset optimaly/uniformly with regards to distribution of Y (Phenos = Drug Resistance) values but also with regards to distributions of repeated measurements (repeated Y Phenos for a the sample sample; required to evaluate Rule 4 for minimal level of accuracy) and some putative explainative variables (X1, X2,X3...). And of course NGYX I.C. checks how good this is performed by the end. No way to start with biased subsets! Example in picture below: Y (Phenos) for drug ABaCavir / ABC. Pearson Correlation between the full dataset and the subset #1 at 0.998.
So to summarize:
We always consider that validation should be performed on a subset of unseen Data equal in size to what is used to develop / test the Methodology. 10 Fold or Leave-One-Out are not used.
While this may be enough (50% vs. 50%) to capture e.g. the confidence on HIV1 Drug Resistance predictions (T Test with 1 degree of freedom), NGYX I.C. splits the dataset in 4 parts (A,B,C & D; accordingly to principles of "uniformization" stated here over) and uses the 6 combinations "to learn". Then the 6 combinations are evaluated with respect of the " 50% seen / 50% unseen).
Not pictured out here over but we also have some rules about variables (e.g. a specific mutation) prevalences:
To incorporate a variable (considered on its own) it should be present in all 4 groups at least 3 times. And for a group of e.g. 100 entries also at least 3 times absent.
If you want to deal with the next level = interactions (level 2; e.g. mutations A and B) you also need the 4 groups to full fil the same prevalences e.g. for the 4 combinations A+B; A alone, B alone, and none of these A &B. 3 Times and not more then 97 times.
There is one thing missing but it is your job to sort it out: You need to find a way to evaluate Under/over fitting. Using a 95% "usual" threshold is not an option (why 95% and not 989%?). NGYX I.C. has developped a very robust methodology for this purpose. But we would be very interested to see something else...
2. Prediction Methodology Performance.
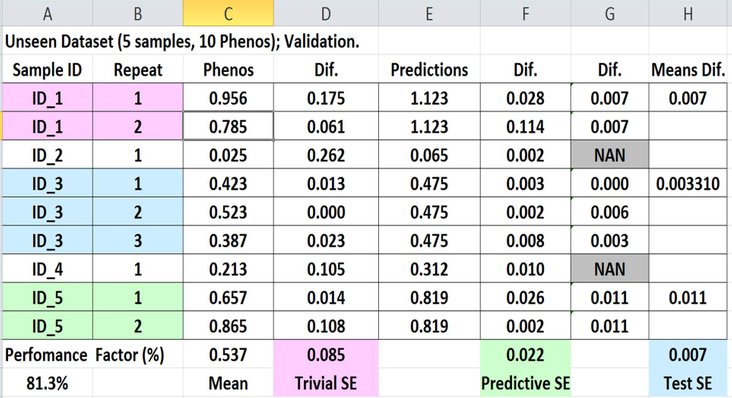
NGYX I.C. has selected its own way to evaluate prediction system performance. We call it the "Performance Factor" (PF).
This PF uses the following formula:
PF = (Trivial SE - Predictive SE) / (Trivial SE - Test SE).
The picture below shows more about the way the PFs are calculated.
AND NGYX I.C. apllies the following rules:
A. New System (no reference, no previous version validated using NGYX I.C. methodology).
In such a case, the median and the mean of the 6 validation PF (remember NGYX I.C. approach) should reach 50%. Means 50% of the way to a optimal predictive system is done.
B. Cases where a NGYX I.C. already approved versions exist.
Rule stipulated in point A. applies. But on the top the new prediction system candidate for implementation needs to be at least equivalent to the previous one (we use a T Test on the Means of PFs).
ATTENTION: Validation should be performed on the same (combination of) validation subsets. This may give a + to the new system that incorporate probably more data in Training/Test step. But that's the point if you want to capture an evolving situation (e.g. appearence of emerging resistance mutations in HIV1).
Now you know all about the Datasets / and Subsets and it is up to you to start looking at these e.g. with the Subsets #1 that are FREE! Click HERE!
II. NGYX I.C. Challenge.
Principe is very easy: Using for all 22 Drugs the FREE Subsets #1 you will use and apply a unique methodology with respect to rules 1 to 6, validate it as described and if you succeed generationg for all Performance Factors reaching 50% you will be qualified and the 3 best will receive for FREE the subsets #25 for eaxh of the 22 FDA approved drugs.
You will have to demonstrate that it makes it also on this second subset. And if so, will will have a more in depth discussion / proposal with/for you.
If you believe you have it, submit your report / documation as attachment to Challenge@NGYX.EU.