NGYX I.C.
WEB Content
Correlated Genos/Phenos Subsets.
EFaVirenz / EFV / R overview.
Remark: Some data relate to Subset#1 (Free) used as an example (1 out of 25). You can access an INFO file with Raw data to analyse and complete the picture. Click HERE.
If you want to catch the Subset#1 Dataset (or even more) Click HERE.
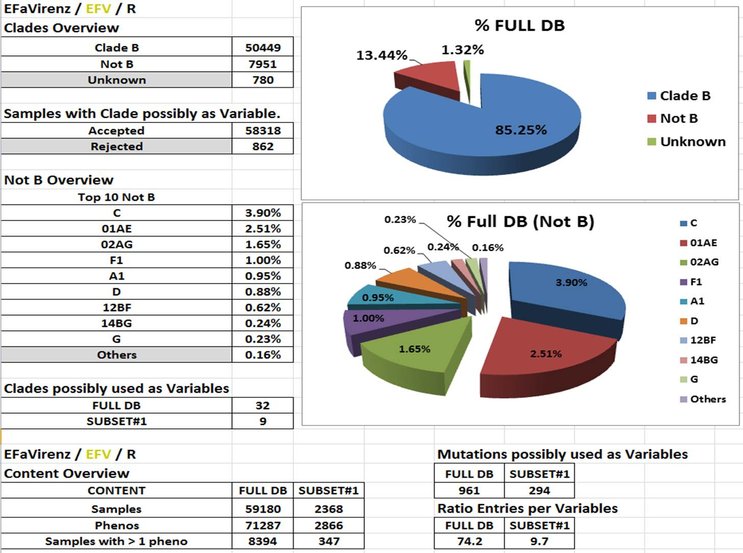
I. G/Ps Content and Clades Prevalance.
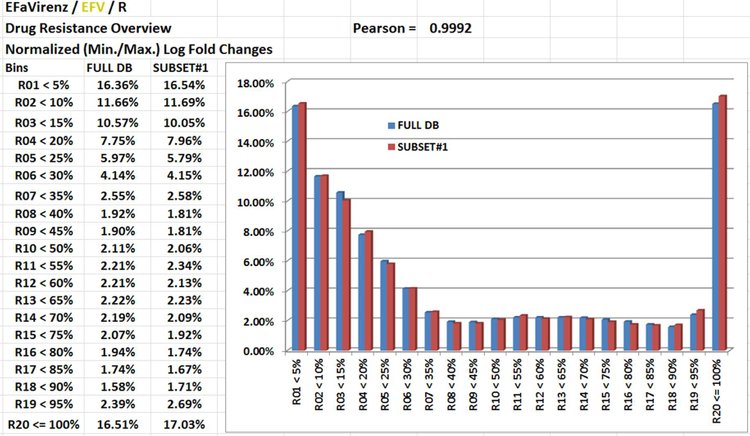
II. Drug Resistance accross G/Ps.
NGYX I.C. Comment(s): There are numerous Phenotypic results (various sources) that fail to be discriminative at both Sensitive/Resistance sides (numerous measurements are under/over the lower/upper limit of the detection range). Definitively EFV would have been beneficial to have data from an extended Test Dynamic Range.
III. Additional Remarks.
A. EFV is rated at 65% on the NGYX I.C.Scale of Drug Resistance Interest (DRI). This means that each couple of G/P is valuable maximally for 0.65 Euros. If you want to know more about DRI Click HERE.
B. Combining subsets will impact the number of variables (mutations / clades) that NGIX I.C. classify as "ACCEPTED". T0 know more about this Click HERE.